
GENETICS AND BIOINFORMATICS
In this issue of the newsletter, we highlight the research being carried out at the Department of Genetics and Bioinformatics, led by Prof. Fahd Al-Mulla, at DDI.
NEWS AND VIEWS
Explore the most recent advances in the world of diabetes and related complications published in the highest impact journals
How Large Is The Repertoire Of T2DM Susceptibility Gene Loci?
Published on 01/08/2019
Genome-wide efforts to delineate the gene loci that are susceptible to type 2 diabetes began to take shape around the year 2000. The study cohorts largely represented people of European descent. Few studies were conducted in other continental populations which invariably led to identification of additional novel risk loci. The count of identified risk gene loci was around 116 until a couple of years back (Figure 3).
Despite many years of global genetics research on type 2 diabetes, it is not yet clear whether its total genetic architecture is deciphered comprehensively. Not every identified gene locus has been dissected to single-variant resolution. In this context, the recent works of Mahajan et al.4 that combined data from 32 European-descent GWAS consisting of about 0.9 million individuals, imputed to high-density reference panels, replicated the previously identified 116 gene loci and further identified an additional 135 genes taking the total count of T2D susceptibility gene to 243 with 403 independent (genotype-phenotype) association signals. This study claims to provide the most up to date and exhaustive coverage of the genetic contribution to type 2 diabetes highlighting on locus discovery, causal variant resolution and mechanistic insight. As a next step, it would be interesting to see how many of these association signals are replicable in ethnic populations such as those of Arabs.
The limitation of global studies conducted so far is that they focus mostly on European individuals, with very less or negligible representation from populations such as the Middle East. In our research here at DDI, we are trying to close this gap, by extensively genotyping, sequencing and collecting available data of individuals from Arab populations.
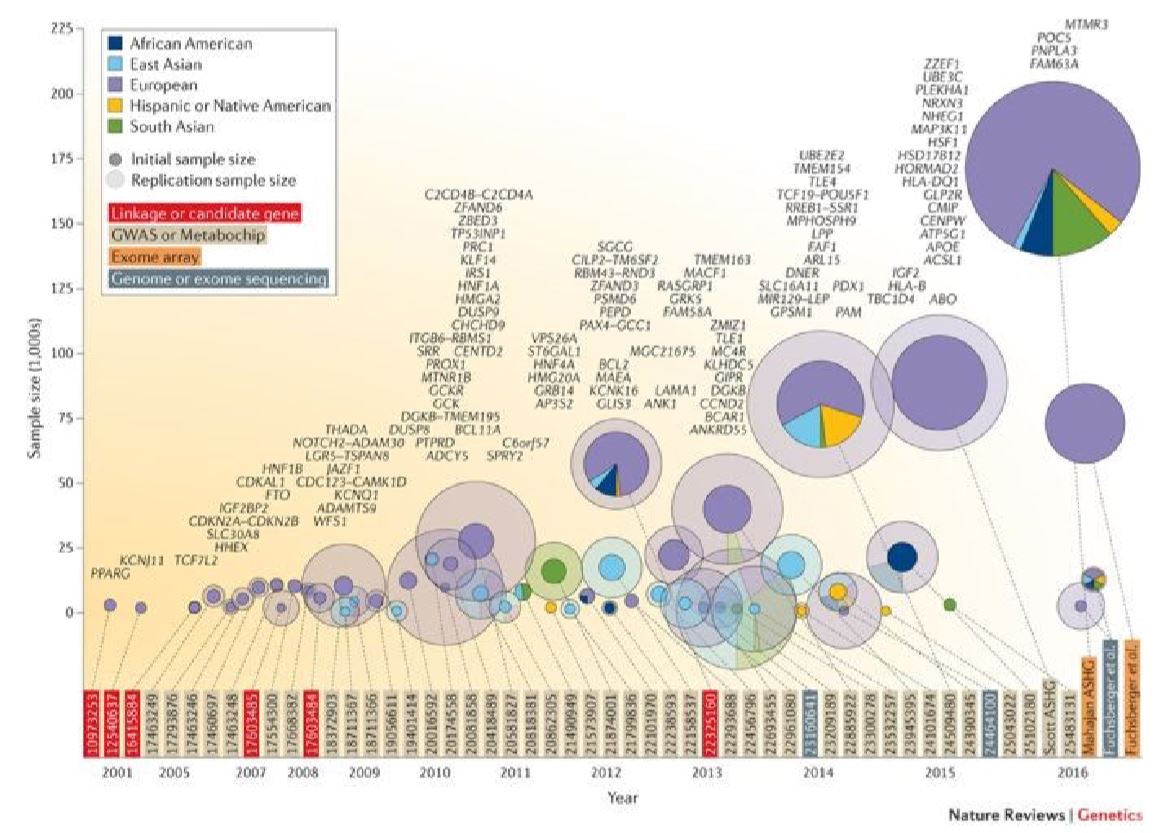
Figure 3: Illustration of relative contributions from global populations to the repertoire of risk loci for type 2 diabetes. Figure is reproduced from the journal of Nature Reviews. Over the years, genome-wide association studies (GWAS) on type 2 diabetes mellitus (T2D), which typically consist of a two-stage discovery and replication study design, have increased in sample size and diversity. The x-axis shows the year of publication, whereas the y-axis shows discovery sample size. The inner (darker) circles are also scaled in proportion to discovery sample size, whereas the outer (lighter) circles are scaled in proportion to total (discovery + replication) sample size. Circles are colored according to ethnic composition of the sample set: African American (dark blue), East Asian (light blue), European (purple), Hispanic or Native American (yellow) or South Asian (green). PubMed identifiers (or first author name) for each study are shown at the base of the figure and connected to the corresponding circle with a dotted line. Identifiers are colored according to the technology used in the study: linkage or candidate gene studies (red), GWAS or Metabochip (beige), exome array (orange) or sequencing (dark grey). Additionally, T2D loci are listed directly above the first reporting study. For example, the biggest circle on the top right-hand corner, shows a large study which identified T2D genetic variants mostly in European samples (purple) and from four gene loci.
4, Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. (2018) 50:1505–13. doi: 10.1038/s41588-018-0241-6


