
BIOCHEMISTRY AND MOLECULAR BIOLOGY DEPARTMENT
In this issue of the newsletter, we will be highlighting the research being carried out by the Biochemistry and Molecular Biology Department at DDI, led by Dr. Jehad Abubaker.
RESEARCH UPDATE
Find out what is new within the Research Sector at DDI
Epigenetics And Epitranscriptomics
Published on 01/09/2019
While Genetics, the study of gene expression, DNA sequencing and genetic variations, is important and has advanced our understanding of many diseases, the last two decades witnessed a renewal of interest in Epigenetics, the study of heritable changes in gene expression without changes in the DNA sequence, i.e. a change in phenotype (observable characteristics) without a change in genotype.
Epigenetic changes are natural occurrences, they happen when genes are actually in action and they can be influenced by several factors during the life span of a human including age, the environment/lifestyle, as well as disease state. Furthermore, epigenetic changes are reversible.
Nessa Carey’s book on Epigenetics Revolution presented a great analogy for Epigenetics. “If you think of the human lifespan as a very long movie, the cells would be the actors and actresses, essential units that make up the movie. DNA, in turn, would be the script, the instructions for all the participants of the movie to perform their roles. Subsequently, the DNA sequence would be the words on the script, and certain blocks of these words that instruct key actions or events to take place would be the genes. The concept of genetics would be like screenwriting. The concept of epigenetics, then, would be like directing. The script can be the same, but the director can choose to eliminate or tweak certain scenes or dialogue, altering the movie for better or worse. After all, Steven Spielberg’s finished product would be drastically different than Woody Allen’s for the same movie script, wouldn’t it?”
The three well-known systems of Epigenetic modifications are DNA methylation, histone modification and noncoding RNA–mediated regulation. Within the field of Molecular Biology, the study of biochemical modifications of RNA is called Epitranscriptomics.
This post-transcriptional gene expression regulation of RNA is critical because it eventually controls protein production. RNA trafficking, translation efficiency, and stability are all controlled at the transcript level. Many nucleotide modifications have been described in diverse types of RNA molecules and every position of pyrimidine and purine rings can be post-transcriptionally modified (Dominissini, 2014). However, methylation remains the most prevalent form of modification. More specifically, N6-methyladenosine (m6A) modifications, which is the addition of a methyl group to adenosine, is the most common RNA modification (Yang, Hsu, Chen, & Yang, 2018). Detailed studies of m6A functions began around 2012, when transcriptome-wide profiling of m6A became possible through antibody-based immunoprecipitation followed by high-throughput sequencing.
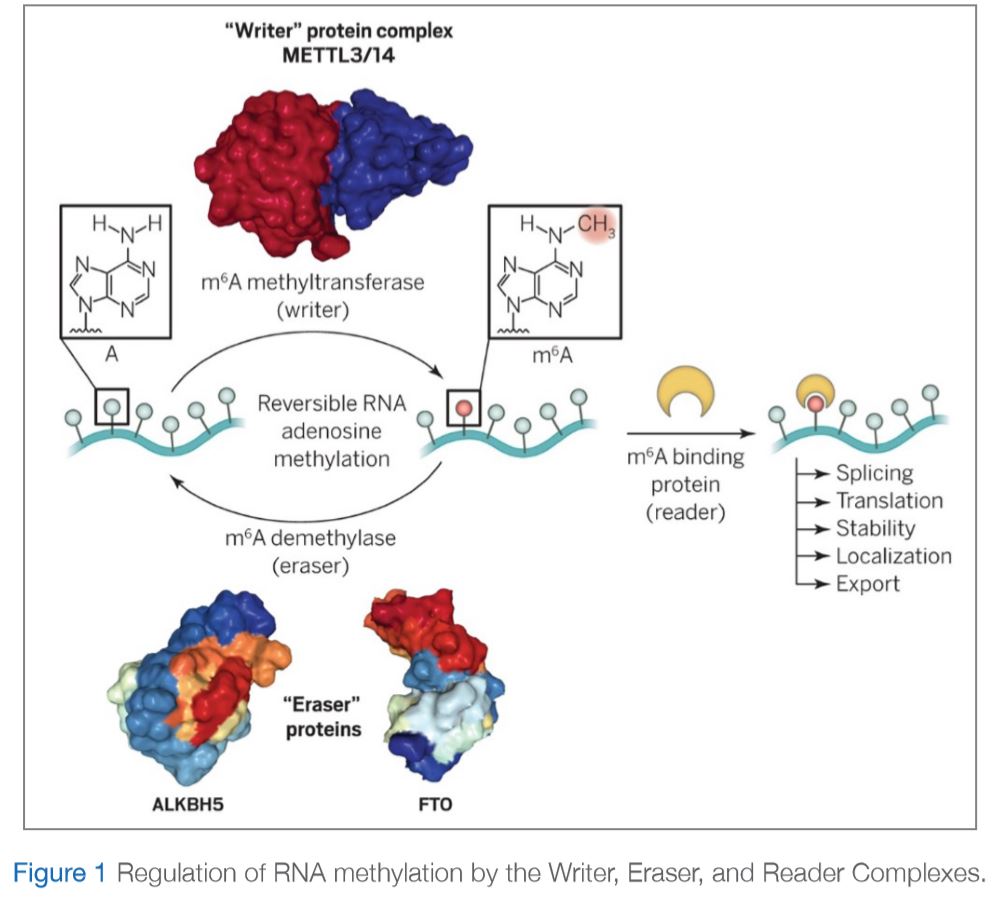
There are three important sets of proteins that mediate this modification as illustrated in Figure 1:
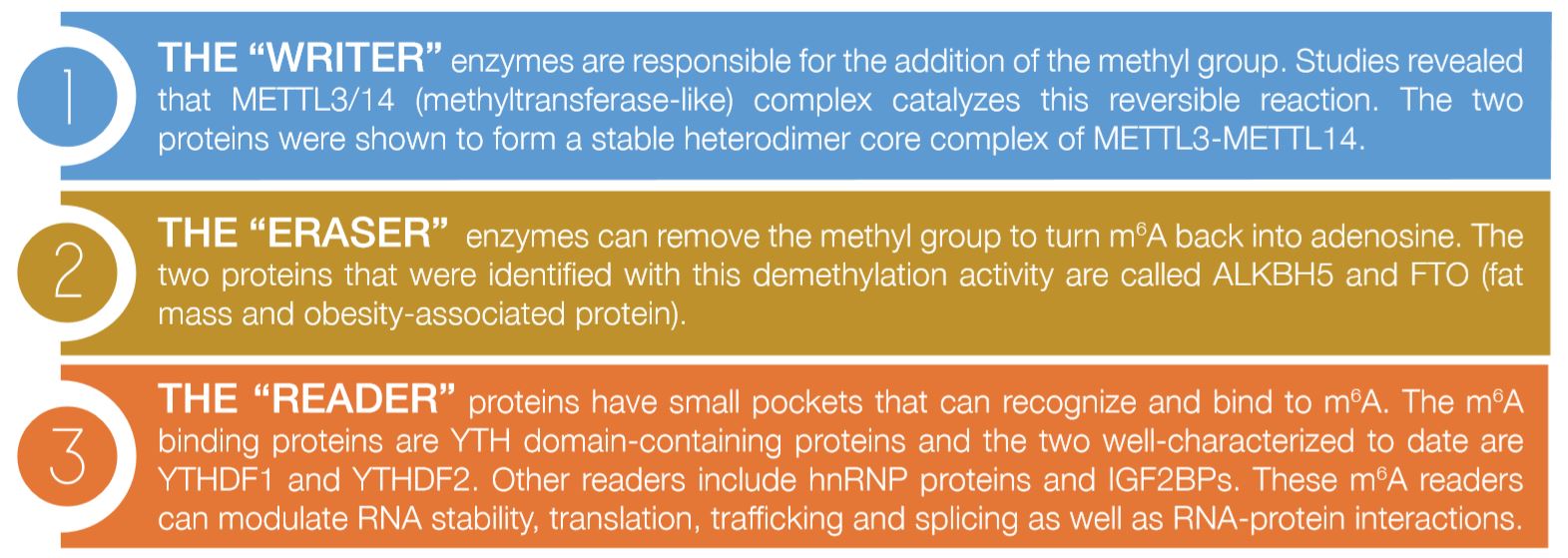
The importance of such RNA modifications is well-known and very evident; however, their mode of action is still unclear. In addition, Epigenetics and Epitranscriptomics have been classically associated with cancer, however, recent studies are exploring their role in different diseases. The Biochemistry and Molecular Biology department at DDI has developed many projects to investigate the role of noncoding RNA, specifically microRNA, in diabetes, obesity and metabolic diseases.
REFERENCES:
1. Abu-Farha, M., P. Cherian, I. Al-Khairi, R. Nizam, A. Alkandari, H. Arefanian, J. Tuomilehto, F. Al-Mulla and J. Abubaker (2019). “Reduced miR-181d level in obesity and its role in lipid metabolism via regulation of ANGPTL3.” Scientific Reports 9(1): 11866.
2. De Jesus, D. F., Z. Zhang, S. Kahraman, N. K. Brown, M. Chen, J. Hu, M. K. Gupta, C. He and R. N. Kulkarni (2019). “m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes.” Nature Metabolism 1(8): 765-774.
3. Dominissini, D. (2014). “Roadmap to the epitranscriptome.” Science 346(6214): 1192.
4. Yang, Y., P. J. Hsu, Y. S. Chen and Y. G. Yang (2018). “Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism.” Cell Res 28(6): 616-624.


