
GENETICS AND BIOINFORMATICS
In this issue of the newsletter, we highlight the research being carried out at the Department of Genetics and Bioinformatics, led by Prof. Fahd Al-Mulla, at DDI.
RESEARCH HIGHLIGHT
Discover the latest research being carried out at DDI
Genetic Variability In Protein-coding Regions Of The Human Genomes From Kuwait
Published on 01/08/2019
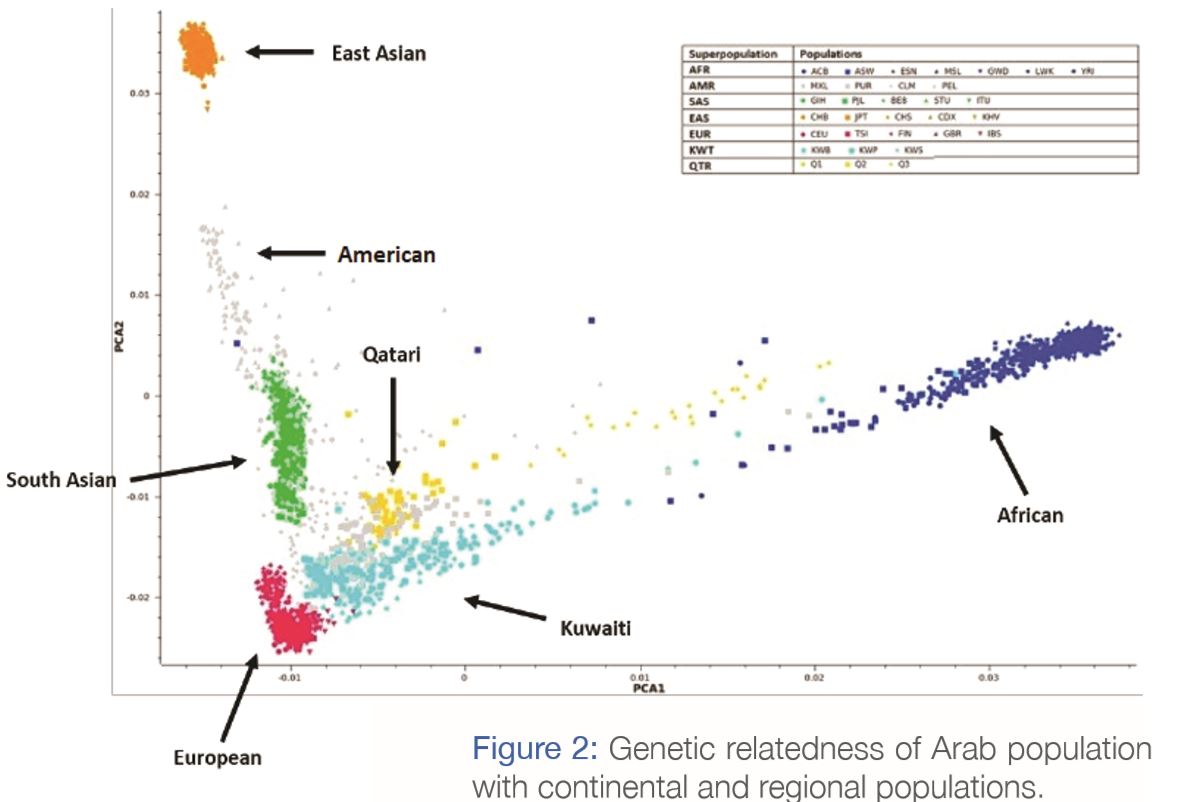
A typical human genome comprises around 3 billion letters (nucleotide bases); however, only a tiny fraction (~1%) of the bases constitute DNA regions that code for proteins. Genetic variability in such protein-coding DNA regions (forming what is called ‘exome’ as opposed to ‘genome’) determines largely the variations in phenotype traits (observable traits such as eye color and blood type) among individuals. Differences in genetic variability in the protein-coding DNA regions may explain population-level differences in the prevalence of both rare genetic disorders and common disorders, and in the efficacy of medications. Major studies on such genetic variability focused primarily on the Caucasian population; as such, capturing the extent of genetic variation in the Arab population is poorly represented in global studies. A recent study1 carried out in Dasman Diabetes Institute sequenced the whole exome of around 500 individuals from Kuwait and presented a “Arabian Exome Variation” database. This allowed characterization of frequent variants related to the onset of common diseases, and to the pharmacogenomics of common disorders such as diabetes & hypertension.
The study identified genetic similarities among Kuwaiti population, Middle Eastern, European and Ashkenazi Jewish populations (Figure 2). The figure shows the distribution of Kuwaiti individuals (in light blue) alongside individuals from global populations. Basically, it describes how individuals from different populations relate to each other genetically, if they were to be plotted on a world genetic map. The figure tells us that the Kuwaitis are genetically very similar to Qataris (in yellow), with a few individuals (of nomadic Bedouin ancestry) showing close affinity to African individuals (dark blue). Among all the populations, we find that the Americans are the most diverse (shown in grey). On the other hand, the East Asians form a well-defined isolated group on the top-left corner (in orange).
This study is a significant addition to regional and global data resources on human exome variability; however, a wide range of similar studies in the region are warranted to support genomic discoveries in medical and population genetics at the regional level.
1, Sumi Elsa John, Dinu Antony, Muthukrishnan Eaaswarkhanth, Prashantha Hebbar, Arshad Mohamed Channanath, Daisy Thomas, Sriraman Devarajan, Jaakko Tuomilehto, Fahd Al-Mulla, Osama Alsmadi, Thangavel Alphonse Thanaraj* (2018). Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Scientific Reports volume 8, Article number: 16583 (2018). DOI:10.1038/s41598-018-34815-8.
GENETIC VARIABILITY IN PROTEIN-CODING REGIONS OF THE HUMAN GENOMES FROM KUWAIT2,3
Global genome-wide association studies (GWAS) have established several genome variants as risk variants for metabolic disorders such as obesity and diabetes. These global studies are largely based on populations of European descent. Ethnic populations such as those of the Arabian Peninsula are not included in these global studies. Genetics research in DDI has demonstrated that the Arab population can contribute to the global catalog of risk variants for metabolic disorders. A recent study2,and previous publications from the institute reporting results of genome-wide association studies, have identified around 30 novel risk loci3 relating to traits associated with obesity, diabetes and dyslipidemia. It is often the case that the gene regions harboring these novel association signals also harbor runs of homozygosity – a continuous stretch of genome region that are homozygous i.e. both the copies of DNA, that every individual possesses, contains the same allele (of the four building blocks of DNA namely T,C,A,G) at the constituent nucleotide position; runs of such homozygous region are hallmark of inbreeding and endogamy seen in the population. Overall, our studies have augmented international efforts that identify the basis for genetic regulation of metabolic traits.
2, Prashantha Hebbar, Rasheeba Nizam, Motasem Melhem, Fadi Alkayal, Naser Elkum, Sumi Elsa John, Jaakko Tuomilehto, Osama Alsmadi, Thangavel Alphonse Thanaraj* (2018) Genome-wide association study of genetic markers for lipid traits in the Arab population. J. Lipid Res. 2018 59:(10) 1951-1966. First Published on August 14, 2018, doi:10.1194/jlr.P080218.
3, Prashantha Hebbar, Jehad Abubaker, Mohamed Abu-Farha, Jaakko Tuomilehto, Fahd Al-Mulla, Thangavel Alphonse Thanaraj (2019) A perception on genome-wide genetic analysis of metabolic disorders in Arab population versus global populations. Front Endocrinol (Lausanne). 10: 8. doi: 10.3389/fendo.2019.00008.
MR


